- Provides information on cancer research
- Outstanding and original reviews
- Suitable for researchers and students
Advances in Cancer Research provides invaluable information on the exciting and fast-moving field of cancer research. Here, once again, outstanding and original reviews are presented on a variety of topics. This volume, number 124, covers emerging applications of molecular imaging to oncology, including molecular-genetic imaging, imaging the tumor microenvironment, tracking cells and vaccines in vivo, and more. Provides information on cancer research Outstanding and original reviews Suitable for researchers and students
Front Cover 1
Emerging Applications of Molecular Imaging to Oncology 4
Copyright 5
Contents 6
Contributors 10
Preface 14
Chapter One: Quantitative Radiology: Applications to Oncology 16
1. Introduction 16
2. Radiological Characterization of Tumors 17
2.1. Computed tomography 18
2.1.1. Structural (routine) CT 18
2.1.2. CT perfusion 18
2.1.3. Dual-energy CT 20
2.2. Magnetic resonance 21
2.2.1. Structural (routine) MR 21
2.2.2. MR spectroscopy and hyperpolarization 21
2.2.3. MR perfusion 24
2.2.4. Diffusion-weighted imaging 26
2.2.5. Diffusion tensor imaging 27
2.3. Positron emission tomography 28
3. Quantitative Radiology 30
3.1. Image analysis 30
3.1.1. Manual segmentation 30
3.1.2. Automated segmentation 32
3.1.3. Registration 34
3.2. Evaluation 35
3.3. Integration 36
4. Future Directions 36
5. Conclusion 38
References 38
Chapter Two: The Intricate Role of CXCR4 in Cancer 46
1. Introduction 47
2. CXCR4/CXCL12 Signaling 48
3. Expression and Physiological Functions of the CXCR4/CXCL12 Axis 50
4. Role of CXCR4 in Cancer 52
4.1. Leukemia 54
4.2. Multiple myeloma 55
4.3. Breast cancer 56
4.4. Prostate cancer 57
4.5. Ovarian cancer 58
4.6. Lung cancer 59
4.7. Gastrointestinal cancers 61
4.8. Renal cell carcinoma 64
4.9. Melanoma 65
4.10. Brain tumors 66
4.11. Soft tissue sarcomas 67
5. CXCR4 Antagonists as Therapeutic and Imaging Agents 67
6. Peptides and Peptidomimetics 70
6.1. CXCL12-based peptides 70
6.2. Synthetic peptide CXCR4 antagonists 71
6.3. Small cyclic peptide analogues 74
6.4. Antibodies against CXCR4 75
6.5. LMW CXCR4 antagonists 76
7. Conclusion 79
Acknowledgments 79
References 79
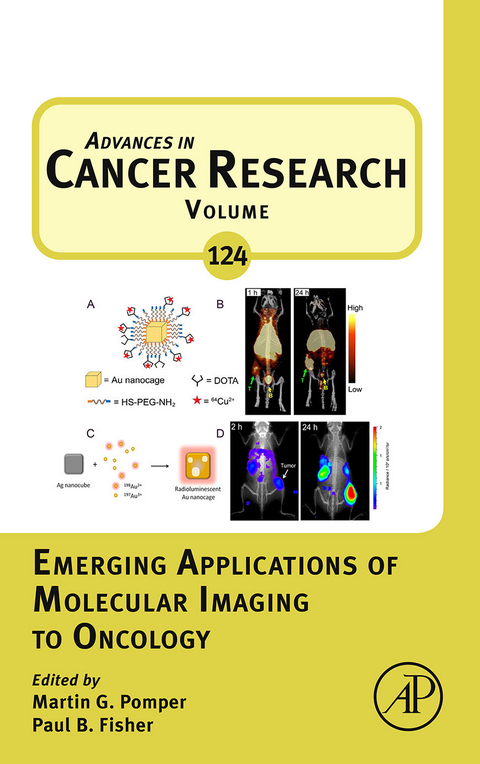
Chapter Three: Recent Advances in Nanoparticle-Based Nuclear Imaging of Cancers 98
1. Introduction 99
2. Lipid-Based Nanoparticles 104
3. Dendrimers 112
4. Polymers 113
5. Quantum Dots 115
6. Iron Oxide Nanoparticles 117
7. Gold Nanoparticles 123
8. Carbon Nanotubes 125
9. Silica-Based Nanoparticles 127
10. Conclusion 132
References 132
Chapter Four: Molecular-Genetic Imaging of Cancer 146
1. Introduction 147
2. Promoters 148
3. Reporters 154
4. Signal Enhancement of Reporters 159
4.1. Enhancers 160
4.2. Two-step transcriptional amplification 161
4.2.1. Bidirectional TSTA 161
4.2.2. Lentivirus-TSTA 163
4.2.3. Adeno-TSTA 164
4.2.4. Advanced TSTA system 164
4.2.5. Replacing components of the TSTA 164
4.2.6. Titratable TSTA 165
4.2.7. Dual TSTA 165
4.2.8. TSTA for imaging cellular differentiation 165
4.3. Codon optimization 166
4.4. Posttranscriptional regulatory elements 166
4.5. Synthetic super promoter 167
4.6. Introducing introns 167
5. Prolonged Expression of Reporters 168
6. Machinery for Gene Delivery 169
6.1. Cationic polymers (polyplexes) 170
6.2. Positively charged lipids (lipoplexes) 170
6.3. Nanoparticles (nanoplexes) 171
7. Size and Immunogenicity 172
8. Concluding Remarks 174
Acknowledgments 175
References 175
Chapter Five: Real-Time Fluorescence Image-Guided Oncologic Surgery 186
1. Introduction 187
1.1. Need for real-time image-guided surgery 188
1.2. Current methods available for image-guided surgery 190
1.3. Optical methods amenable to image-guided surgery 191
2. Fluorescence Imaging Systems for Intraoperative Procedures 193
2.1. Fluorescence sensor parameters 193
2.1.1. Quantum efficiency of a photodiode 194
2.1.2. Signal-to-noise ratio of an imaging sensor 194
2.1.3. Electrical and optical crosstalk 197
2.1.4. Transmission and optical density of excitation and emission filters 199
2.1.5. Overall SNR and contrast ratio of fluorescence signal 199
2.2. Optical design parameters 201
2.2.1. Lens and filter strategy 201
2.2.2. Illumination design 202
3. Current Intraoperative Optical Image Guidance Systems 205
4. Fluorescent Agents Used in Image-Guided Surgery 207
4.1. Endogenous fluorophores 208
4.2. Exogenous fluorescent agents 210
4.2.1. Fluorescein 210
4.2.2. Methylene blue 211
4.2.3. 5-Aminolevulinic acid 212
4.2.4. Indocyanine green 212
5. Clinical Applications of Fluorescence Image-Guided Surgery 213
5.1. Sentinel lymph node mapping 213
5.2. Tumor imaging 215
6. Future Directions 216
7. Concluding Remarks 217
References 218
Chapter Six: Cerenkov Imaging 228
1. Introduction 229
2. Cerenkov Radiation Physics (Simplified) 229
2.1. Dependence of CL on the refractive index of the medium 230
2.2. CL from a-particles 231
2.3. Conical wave front of Cerenkov light 232
2.4. Spectral characteristics of Cerenkov 233
2.5. Light intensity and spatial distribution 233
2.6. CL in tissue 236
3. Application of Cerenkov in Biological Sciences: CLI 237
3.1. CL from medical radiotracers 237
3.2. Instrumentation for CLI 238
3.3. Cerenkov luminescence tomography 239
3.4. Clinical Cerenkov imaging 241
3.5. Intraoperative Cerenkov imaging 242
3.6. Cerenkov to improve positron emission tomography 244
3.7. Cerenkov 2.0 244
4. Conclusion 246
Acknowledgments 246
References 246
Chapter Seven: Molecular Imaging of the Tumor Microenvironment for Precision Medicine and Theranostics 250
1. Introduction 251
2. Imaging and PM/Theranostics of the Physiological Microenvironment 253
2.1. Hypoxia 253
2.2. pH 256
3. The ECM and Its Enzymes 259
4. Endothelial Cells and Tumor Vasculature 261
5. Lymphatic Endothelial Cells, Lymphatics, and Interstitial Pressure 263
6. Stromal Components of the TME and Their Role in PM 264
7. Intraoperative Optical Imaging 266
8. Concluding Remarks 267
Acknowledgments 267
References 267
Chapter Eight: Tracking Cellular and Immune Therapies in Cancer 272
1. Introduction 273
1.1. History of cancer immunotherapy and passive versus active immunity 274
1.2. Immune cell subsets, the immunosuppressive microenvironment, and checkpoint inhibitors 275
1.3. Cellular therapies-Dendritic cell vaccines and CAR-T cells 277
1.4. Shortfalls of anatomic imaging for immunotherapies 278
2. Molecular Imaging Approaches to Cancer Immunotherapy 280
2.1. Approaches toward imaging the immune system 280
2.2. Applicable imaging modalities 283
3. Radionuclide Methods in the Preclinical and Clinical Settings 285
3.1. Direct labeling methods 285
3.2. Indirect imaging with reporter genes 286
3.3. Enzyme-based strategies 287
3.4. Receptor-based strategies 289
3.5. Transporter-based strategies 290
4. MRI Methods in the Preclinical and Clinical Settings 291
4.1. Types of MRI contrast agents 291
4.2. SPIO imaging 293
4.3. 19F MRI using perfluorocarbons 295
5. Opportunities for Improvements and Future Directions 297
5.1. Imaging the tumor immune environment prior to immune therapy 297
5.2. Imaging immune checkpoints 298
5.2.1. Cytotoxic T-lymphocyte-associated antigen 4 299
5.2.2. Programmed death 1 300
5.3. Opportunities for predicting and assessing immune responses 301
5.4. In vivo cell labeling progress to date 302
5.5. Imaging cell state with reporter genes 303
6. Conclusions 303
References 304
Chapter Nine: Developing MR Probes for Molecular Imaging 312
1. General Overview 313
2. T1, T2, T2* Relaxivity-Based Agents 315
3. CEST Probes: Multiple Labeling Frequencies 317
3.1. Diamagnetic CEST probes 320
3.2. Paramagnetic CEST probes 322
3.3. Nanoparticle-based CEST probes 323
4. 19F Probes: Hot-Spot Imaging 323
4.1. 19F-containing metal complexes 324
4.2. 19F-containing nanoemulsions 324
5. Hyperpolarized Imaging Probes 325
5.1. Dynamic nuclear polarization 326
5.2. Parahydrogen-induced polarization 329
5.3. Spin exchange optical pumping 330
References 331
Chapter Ten: Clinical Translation of Molecular Imaging Agents Used in PET Studies of Cancer 345
1. Introduction 345
2. FDG-Lessons Learnt 352
3. Stages to Development of a New Radiotracer 354
4. Translating Deregulated Nature-Identical Biochemicals 356
4.1. Choline metabolism 356
4.2. Fatty acid metabolism 359
4.3. Amino acid metabolism 359
5. Translating Cell Surface and Intracellular Receptors as Predictive Biomarkers 360
5.1. Epidermal growth factor receptor in lung cancer 360
5.2. HER2 in breast cancer 361
5.3. ER signaling in breast cancer 362
5.4. PSMA in prostate cancer 363
6. Translating Probes for Visualization of Life and Death Signals in the Cell 364
6.1. Proliferation 364
6.2. Apoptosis 369
7. Translating Tools to Assess Host-Tumor Microenvironment Interactions 371
7.1. Angiogenesis 371
7.2. Hypoxia imaging 373
8. Translating Labeled Drugs and Drug Analogs 374
9. Conclusion 375
Acknowledgments 375
References 376
Index 390
Color Plate 398
Quantitative Radiology
Applications to Oncology
Edward H. Herskovits1 Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland, Baltimore, Maryland, USA
1 Corresponding author: email address: ehh@ieee.org
Abstract
Oncologists, clinician-scientists, and basic scientists collect computed tomography, magnetic resonance, and positron emission tomography images in the process of caring for patients, managing clinical trials, and investigating cancer biology. As we have developed more sophisticated means for noninvasively delineating and characterizing neoplasms, these image data have come to play a central role in oncology. In parallel, the increasing complexity and volume of these data have necessitated the development of quantitative methods for assessing tumor burden, and by proxy, disease-free survival.
Keywords
Quantitative radiology
Oncology
CT
MR
Molecular imaging
Image segmentation
Image registration
Data mining
1 Introduction
Oncologists, clinician-scientists, and basic scientists collect a plethora of data in the process of caring for patients, managing clinical trials, and investigating cancer biology. As we have developed more sophisticated means for noninvasively delineating and interrogating neoplasms, the resulting image data have come to play a central role in oncology. To understand the current impact and long-term promise of radiology with respect to oncology, it may help to characterize the nature of the information sought as we diagnose and treat cancer patients.
The ultimate goal of patient care in oncology is to maximize disease-free survival (DFS)—or, barring that, progression-free survival (PFS)—while minimizing the morbidity of treatment (i.e., to maximize quality-adjusted life years). Ignoring intercurrent illnesses and treatment morbidity for the sake of this discussion, we take PFS to be a function of tumor burden, which can be decomposed into two independent factors: the number of tumor cells and the malignant potential of each cell. For many years, the former—extent—was determined via exploratory surgery and summarized as tumor stage, and the latter—grade—was determined by pathologists from what was hoped to be a biologically representative sample obtained during this operation. Advances in radiology first became evident with respect to staging, for the simple reason that it is much easier to generate images that show macroscopic groups of cells than it is to generate images that show how these cells are likely to behave. Only in the last decade has radiology begun to offer information regarding tumor biology, and such information still pales in comparison with that obtained from histopathology and genetic analysis.
In parallel with the increasing complexity of image data, there has been steady progress in the quantification of these data. Although clinical radiology reports are unfortunately replete with verbiage such as “large mass in the right frontal lobe,” researchers have begun to deliver on the promise of computer-based methods for quantification of tumor extent and have also developed quantitative or semiquantitative methods for characterizing tumor biology. The premise underlying such efforts is that quantitative—rather than qualitative—indications of tumor extent and biology render more precise prediction of DFS, thereby promising superior patient care and assessment of therapy. Herein I explore the arc of radiology's contributions to oncology, both in terms of the information provided and efforts to quantify this information, with the expectation that such exploration will shed light on future developments in oncology research and practice.
2 Radiological Characterization of Tumors
The advent of computed tomography (CT) revolutionized the staging of solid tumors; since then, the quality and range of information provided to oncologists via noninvasive radiological examinations have steadily increased. The vast majority of this information relates to tumor extent; however, magnetic resonance (MR), positron emission tomography (PET), and newer modalities have offered progressively more detailed information about tumor physiology (Fass, 2008). Despite improvements in these modalities, there remain significant problems. For example, it is well known that, even with a combination of advanced MR sequences such as DTI and perfusion, we cannot accurately delineate the extent of infiltrative tumors, such as glioblastoma. Although there has been a striking expansion of research modalities for characterizing tumors (Budde & Frank, 2009; Cai & Chen, 2008; Desar et al., 2009; Fass, 2008; O'Connor et al., 2008; Pfannenberg et al., 2007; van der Meel et al., 2010; Weissleder & Pittet, 2008), we focus here on those most widely applied in clinical research and practice: CT, MR, and PET.
2.1 Computed tomography
The principal forms of CT used in oncology are structural (routine) CT, CT perfusion, and dual-energy CT (DECT).
2.1.1 Structural (routine) CT
CT, usually following intravenous iodinated-contrast administration, has been the workhorse of oncologists and researchers seeking to stage tumors and determine response to therapy. Relative to MR, PET, and other molecular imaging techniques, CT is inexpensive, fast, applicable throughout the body, and widely available, all of which are critical features of a modality that would be used to establish internationally accepted response criteria for a broad range of neoplasms. For many solid tumors—including some of the most common, such as lung cancer and gastrointestinal malignancies—the contrast between tumor and adjacent normal structures (i.e., tissue contrast) is sufficient to support delineation of lesions (i.e., to estimate stage or tumor burden). With the advent of helical CT (Van Hoe et al., 1997) and multidetector CT, spatial resolution (particularly in the z-axis) increased, allowing characterization of ever-smaller lesions.
2.1.2 CT perfusion
Although CT provides excellent anatomic information for most tumors, it provides little physiologic information about tumors. CT perfusion, in which intravenous contrast is administered as a bolus, and voxel-wise time-attenuation curves are computed from repeated scans (including a baseline noncontrast scan), is one of the most common means for obtaining information beyond precontrast or postcontrast attenuation values. The widespread availability, first of helical scanners, and subsequently of multidetector scanners, has promoted CT perfusion from a research tool into a commonly used clinical tool, with applications across organ systems and disease categories (Miles & Griffiths, 2003).
There are several categories of mathematical models that have been used to inform the calculation of perfusion parameters from the time-attenuation curve, with varying assumptions about the interactions among the contrast material (e.g., diffusibility; bolus contour), the patient's physical state (e.g., cardiac output), and tissue characteristics (e.g., collateral flow; differences between capillary and arterial hematocrit), among others. Virtually all perfusion analysis models ultimately invoke the Fick principle, which codifies conservation of mass—in this case, blood—in the perfusion model. The Fick principle models perfusion with a single (arterial) input that supplies a volume of tissue, which in turn drains into a single (venous) output. Under this model, all contrast must be either in an artery, perfusing tissue, or in a vein.
Two major groups of methods for perfusion analysis are those based on deconvolution, and everything else. Deconvolution methods are considered to be more accurate than alternative approaches, but are also more complex. Methods that do not employ deconvolution, such as the maximum-slope method, often rely on simplifying assumptions, such as the assumption that no venous outflow has occurred during the time interval of interest; although such assumptions are clearly not valid in most CT perfusion acquisitions, they may introduce only minimal parameter estimation errors. Deconvolution approaches assume that the concentration of the contrast agent in tissue is a linear function of flow to the tissue and the convolution of the arterial input function and tissue-specific characteristics (Nabavi et al., 1999); there are fewer simplifying assumptions than in nondeconvolution approaches. Mathematical deconvolution methods, such as singular value decomposition (Kudo et al., 2009; Ostergaard, Weisskoff, Chesler, Gyldensted, & Rosen, 1996), yield blood flow (BF), blood volume (BV), and other tissue-specific parameters. An additional parameter that is commonly employed in clinical practice is mean transit time (MTT), which is computed from flow and volume via the relation BF × MTT = BV. Perfusion is expressed as BF per 100 g of tissue, which can be computed from BF, BV, and a tissue-density conversion factor.
Although CT perfusion has been utilized primarily in evaluating stroke patients, it has also found an important role in the evaluation of neoplasms. To the extent that neovascularity reflects tumor grade, features derived from CT perfusion will prove prognostically useful, particularly...
| Erscheint lt. Verlag | 1.10.2014 |
|---|---|
| Sprache | englisch |
| Themenwelt | Medizin / Pharmazie ► Medizinische Fachgebiete ► Onkologie |
| Medizin / Pharmazie ► Medizinische Fachgebiete ► Radiologie / Bildgebende Verfahren | |
| Naturwissenschaften ► Biologie ► Genetik / Molekularbiologie | |
| Technik | |
| ISBN-10 | 0-12-411634-5 / 0124116345 |
| ISBN-13 | 978-0-12-411634-4 / 9780124116344 |
| Haben Sie eine Frage zum Produkt? |
Größe: 23,8 MB
Kopierschutz: Adobe-DRM
Adobe-DRM ist ein Kopierschutz, der das eBook vor Mißbrauch schützen soll. Dabei wird das eBook bereits beim Download auf Ihre persönliche Adobe-ID autorisiert. Lesen können Sie das eBook dann nur auf den Geräten, welche ebenfalls auf Ihre Adobe-ID registriert sind.
Details zum Adobe-DRM
Dateiformat: PDF (Portable Document Format)
Mit einem festen Seitenlayout eignet sich die PDF besonders für Fachbücher mit Spalten, Tabellen und Abbildungen. Eine PDF kann auf fast allen Geräten angezeigt werden, ist aber für kleine Displays (Smartphone, eReader) nur eingeschränkt geeignet.
Systemvoraussetzungen:
PC/Mac: Mit einem PC oder Mac können Sie dieses eBook lesen. Sie benötigen eine
eReader: Dieses eBook kann mit (fast) allen eBook-Readern gelesen werden. Mit dem amazon-Kindle ist es aber nicht kompatibel.
Smartphone/Tablet: Egal ob Apple oder Android, dieses eBook können Sie lesen. Sie benötigen eine
Geräteliste und zusätzliche Hinweise
Buying eBooks from abroad
For tax law reasons we can sell eBooks just within Germany and Switzerland. Regrettably we cannot fulfill eBook-orders from other countries.
Größe: 15,1 MB
Kopierschutz: Adobe-DRM
Adobe-DRM ist ein Kopierschutz, der das eBook vor Mißbrauch schützen soll. Dabei wird das eBook bereits beim Download auf Ihre persönliche Adobe-ID autorisiert. Lesen können Sie das eBook dann nur auf den Geräten, welche ebenfalls auf Ihre Adobe-ID registriert sind.
Details zum Adobe-DRM
Dateiformat: EPUB (Electronic Publication)
EPUB ist ein offener Standard für eBooks und eignet sich besonders zur Darstellung von Belletristik und Sachbüchern. Der Fließtext wird dynamisch an die Display- und Schriftgröße angepasst. Auch für mobile Lesegeräte ist EPUB daher gut geeignet.
Systemvoraussetzungen:
PC/Mac: Mit einem PC oder Mac können Sie dieses eBook lesen. Sie benötigen eine
eReader: Dieses eBook kann mit (fast) allen eBook-Readern gelesen werden. Mit dem amazon-Kindle ist es aber nicht kompatibel.
Smartphone/Tablet: Egal ob Apple oder Android, dieses eBook können Sie lesen. Sie benötigen eine
Geräteliste und zusätzliche Hinweise
Buying eBooks from abroad
For tax law reasons we can sell eBooks just within Germany and Switzerland. Regrettably we cannot fulfill eBook-orders from other countries.
aus dem Bereich